- Description
- Absolute configuration at the α position
- Amino acids as dipolar ions
- Classifications
- Acidic or basic
- Hydrophobic or hydrophilic
- Reactions
- Sulfur linkage for cysteine and cystine
- Peptide linkage: polypeptides and proteins
- Hydrolysis
- Structure
- 1° structure of proteins
- 2° structure of proteins
- 3° structure of proteins; role of proline, cystine, hydrophobic bonding
- 4° structure of proteins (BIO, BC)
- Conformational stability
- Denaturing and folding
- Hydrophobic interactions
- Solvation layer (entropy) (BC)
- Separation techniques
- Isoelectric point
- Electrophoresis
Review:
Central dogma of Biology:
DNA --> RNA --> proteins
The same information is preserved & passed along all three of the above "steps." Ex: you can find the DNA base code of a protein.
DNA and RNA = nucleic acids
proteins = amino acids
- replication - DNA copying itself
- transcription - DNA becomes RNA
- tranSCRIPTion involves a script. Scripts involve letters. You are going from one written letter form to another written letter form (nucleic acid --> nucleic acid)
- translation - RNA becomes protein
- TRANSLATION is like translating languages. You are going from one language <nucleic acid> into an entirely new language <amino acids>
DNA, RNA, & proteins are ALL monomers (a thing that is attached to ONLY two other subunits) that link together and form a linear polymer.
The monomer for DNA is deoxyribonucleic acid.
The monomer for RNA is ribonucleic acid.
The monomer for proteins are amino acids.
Reverse tranSCRIPTion is also a possibility. You can go from RNA to DNA.
An enzyme (reverse transcriptase) is required for this process to occur. Reverse transcriptase will generate a complimentary DNA molecule (cDNA) from an RNA template.
This is usually seen with retroviruses (ex. HIV - goes from RNA to DNA. The DNA can then integrate itself into other cells genetic make-up).
The genetic make-up of an RNA virus is RNA, not DNA. This allows the virus to form a protein immediately using itself as a template (ex. influenza & SARS).
Noncoding RNA (ncRNA) also exists. Noncoding RNA will NOT form a protein. The most common examples of noncoding RNA are tRNA and mRNA, both of which are used to translate messenger RNA into proteins.
Epigenetic's is when you have different phenotypes derived from the same DNA base. An example of this involves cells. Both muscle cells and skin cells have the same genetic material located within their nucleus. However, both of these cells have VERY different functions, even though they share the same exact DNA base. This is because only certain pieces of the DNA base will be TRANSCRIBED for the cells, based on where they are located.
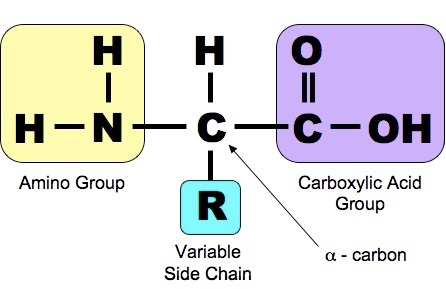
Amino Acids
Amino Acid Structure
Only the L (levorotatory) form of the amino acid is found in the human body!
The alpha carbon is chiral, meaning it will "react" with plane-polarized light.
Light will rotate clockwise if dextro, and counter-clockwise if levo.
D & L and enantiomers of each other.
What is a protein?
A protein is made up of a chain of amino acids, joined together by a peptide bond.A peptide bond forms from the nucleophilic addition/elimination that occurs between the N on an amino acid and the alpha C on an adjacent amino acid.
The peptide bond is resonance stabilized. Because of this, there is a lot of "double bond character" that exists within this molecule (C=O and C=N). Because there is a lot of "double bond character," the peptide bond is rigid. Note: the rest of the molecule is not rigid, just the peptide bond.
This can repeat, forming an endless chain. However, the base pattern will be the same:
Nitrogen - alpha C - carbonyl C
The end of the molecule near the nitrogen is called the N-terminal (amino cap). The opposite end - near the carbonyl C is called the C-terminal. Each group (N-C-C) within the chain is known as a residue.
The peptide bond can break apart (forming individual amino acids) in the following ways:
- hydrolysis (acid hydrolysis & heat): non-specific cleavage of peptide bond
- proteolysis: specific cleavage of peptide bonds
- by using the protein protease, you are able to chose which specific peptide bonds you would like to cleave
- ex. trypsin will cleave the C-terminal end of the amino acids Arg & Lys: N-terminal - Thr - Arg - His - Pro - Lys - Val - C-terminal add trypsin will produce the following segments: Thr - Arg and His - Pro - Lys and Val
Broad Idea (know for the MCAT): the general difference between hydrolysis and proteolysis
Protein Structures (4 levels)
- Primary structure (1 degree): peptide bond/lineage (aa-aa-aa-aa)
- Secondary - backbone: depends on hydrogen bonding; can have either an alpha helix or a beta sheath
- alpha helix: spiral/spring shape, stabilized by H-bonds
- beta-sheath: parallel (N-C to N-C) or anti-parallel (N-C to C-N)

- Tertiary - distant interactions
- depends on: H-bonds, hydrophobic packing (water molecules surround a molecule, forcing all polar bonds to arrive at the surface and all non-polar bonds to gather in the middle), van-der-waal forces, disulfide bridge formation
- these are folds that will eventually refold into a secondary-backbone structure
- Quaternary - multiple polypeptides
- same interactions as tertiary
- can form tetramer (four polypeptides joined together), dimer, trimer, multimer
Visual Overview:
All four of these classifications of structure HAVE to be correct in order to have conformational stability. If even one structure has an error, the protein will be a "denatured protein" - it will not function as it should.
To denature a protein in a lab, change one of the following:
- temperature: shakes bonds between amino acids, resulting in unfolding (affects 2, 3, 4)
- pH: pH can generally be thought of as the addition/subtraction of protons/H. So pH generally affects ionic (charged) bonds. A change in pH will affect all ionic bonds within the molecule (affects 3 and 4)
- chemical: affects H bonds (1, 2, 3, 4)
- enzyme: breaks peptide bonds, allowing absorption by the body
Classification of Amino Acids
Amino acids can be classified based on the differences between the "side" (R) groups.
The "side" (R) group can vary in the following ways: charge, hydrogen bonding ability, acidic/basic
Non-polar: hydrophobic (avoids water) Polar: hydrophilic (loves water)
Within non-polar, you will find two subcategories:
- alkyl (R group is made up of alkyl groups)
- aromatic (R group has an aromatic present)
Within polar, there are three subcategories:
- neutral/polar R groups: R group has an O/S, which loves electrons so it has a negative charge, allowing a positive charge to be distributed throughout the rest of the molecule
- acidic/negatively charged R group: has a carboxylic acid in the R group, making it an excellent H donor
- Note: once the H atom has been donated, aspartic acid becomes aspartate & glutamic acid becomes glutamate
- basic/positively charged R group: has N atoms present. N LOVES protons!
Four unique amino acids
- Histidine: has a pKa of 6.5, which is equivalent to our physiological pH
- Note: pH < pKa: protonated
- pH > pKa: deprotonated
- Because the pKA is equivalent to our physiological pH, histidine will exist in both protonated and deprotonated forms
- Why is this important? Because this "mixture" can be used to stabilize/destabilize a substrate at the active site of a protein
- Proline: alpha helix breakers
- additional bond on the N, so it has a secondary alpha amino group
- Glycine: alpha helix breaker & achiral center
- The R group has two H attached to it, eliminating chirality. This amino acid is not optically active (rest of the amino acids are), so it will not show up when faced with plane polarized light
Now what is an "alpha helix breaker?"
An alpha helix breaker will disrupt a secondary protein structure. Ex: alpha helix (spring-like in shape). Add Gly or Pro? There will be a "kink" on the normally helical structure.
4. Cysteine: disulfide bridge former
- Cysteine has a thiol (-SH group) attached to it
- -SH groups can easily for S-S bonds with other cysteine molecules
- OIL RIG (oxidation is loss and reduction is gain)
- Reducing conditions: cysteine is alone/no disulfide bridge
- Usually exists in the intracellular space of a cell
- (Trick to help you remember) antioxidants are found in the intracellular space of a cell. Anti - OXIDants = anti- OXIDATION
- H is still present
- Oxidizing conditions: cystine exists as a pair/disulfide bridge forms
- Usually found in the extracellular space of a cell
- H has left
- Cysteine vs cystine:
- the additional e is added to the name under reducing conditions, or when the cysteine molecule is "alone."
- cystine occurs under oxidizing conditions - a disulfide bridge is formed
Isoelectric point & Zwitterion (dipolar ions)
When a molecule is isoelectric, it is uncharged. In this case, the molecule in question is an amino acid.
Anything that accepts protons/H is called "basic." Basic compounds generally have a higher pH (14).
Anything that donates protons/H is called "acidic." Acidic compounds generally have a lower pH (1).
A zwitterion is a molecule that is neutral. It has to have both a positive and a negative charge.
In an amino acid, the amino group is basic. It accepts protons, creating a + charge on the N.
On the other hand, the carboxylic acid is acidic. It donates protons to an -OH, creating a - charge on the O.
A zwitterion would have a + on the N in the amino group AND a negative charge on the O in the carboxylic acid group.
To find the isoelectric point (pI) of an amino acid, take the pKa's of all of the functional groups present, add them together, and divide that total by the number of functional groups present.
(pKa + pKb + pKc)/3 = pI
ex. for a general amino acid, carboxylic acid has a pKa of 2. And amino group has a pKa of 9.
(9+2)/2 = 5.5
----------
Sources:
1. www.khanacademy.com
2. www.aamc.org
3. Amino acid general structure: http://study.com/cimages/multimages/16/amino_acid_med.jpeg
4. Dextro vs levorotatory: http://science.uvu.edu/ochem/wp-content/images/D/dlconvention6.png
5. Peptide bond formation: http://alevelnotes.com/content_images/i3_peptide_bond.png
6. Beta sheath (parallel vs antiparallel): https://kimberlybiochemist.files.wordpress.com/2013/02/sheet11.gif
7. Protein structures overview: https://www.umass.edu/molvis/workshop/imgs/protein-structure2.png
8. Amino acid classification: http://usercontent1.hubimg.com/5788430_f520.jpg
9. Disulfide bond formation: http://www.web-books.com/MoBio/Free/images/Ch2A3.gif
MCAT Summary (brief review two weeks prior to exam):
(1A) Amino Acids
- Absolute configuration at the alpha position: R vs S, D vs L
- Amino acids as dipolar ions: Zwitterion: both a + (N on amino group) and a - (O on carboxylic acid) charge present on one amino acid.
- Acidic vs Basic: acidic indicates that a carboxylic acid is present in the R group and the word "acid" is in the name of amino acid. Basic indicates that a N is present in the R group (because N loves H).
- Hydrophobic vs hydrophilic: nonpolar compounds are hydrophobic. The R groups will contain alkyl or aromatic groups. Polar compounds are hydrophilic. The R group will either be neutral (contains O or S), acidic (carboxylic acid), or basic (contains N).
- Sulfur linkage for cysteine and cystine: cysteine is "alone." Cystine consists of two cysteine molecules joined together by a disulfide bridge (S-S bond). Disulfide bridges form within R-groups.
- Peptide bonds: forms between two amino acids. Specifically, a peptide bond forms between the N in the amino group and the adjacent alpha carbon. Water is lost during the process. Occurs through nucleophilic addition/elimination. The molecule is resonance stabilized around the peptide bond, creating a lot of "double bond" character. This makes the peptide bond especially rigid.
- Polypeptides are chains of amino acids (joined via peptide bonds)
- Proteins are multiple polypeptides joined together
- Hydrolysis is the nonspecific cleavage of a peptide bond, creating random amino acid sequences. Proteolysis, on the other hand uses a protein to allow specific cleavage within a polypeptide chain.
(1A) Protein Structures
- primary vs secondary vs tertiary vs quaternary: primary = linear chain of amino acids. secondary = alpha helix or beta sheath (parallel vs anti-parallel) folding of the linear chain. tertiary: further, distant interactions involving the R groups (will eventually become apart of secondary folds), quaternary: multiple polypeptides joined together to form a protein
- primary (peptide bond linkage). secondary (H bonds). tertiary & quaternary (H bonds, van der waals, disulfide bridge, hydrophobic packing)
- proline: alpha helix breaker
- cysteine: disulfide bridge former
- denaturing: all four structures have to be COMPLETELY correct. If all four are correct, the protein will fold correctly. However, if not, the protein will not function as it is supposed to (denature).
- solvation layer: layer surrounding the protein, can sometimes be water
- separation technique: isoelectric point: when the molecule in question (in this case, an amino acid) is uncharged. Can be calculated by taking the pKa's of ALL of the functional groups present, and dividing by n.
- separation technique: protein electrophoresis: separates proteins based on size and electrical charge
Key Vocabulary words (review daily):
(1A) Amino Acids
- alpha carbon
- R vs S
- L vs D
- dipolar ions
- Zwitterion
- isoelectric point (pI)
- acidic vs basic
- non-polar vs polar
- groups found within ALL amino acids (basic groups + variable R groups)
- how to classify amino acids based on the R group present
- non-polar vs polar
- hydrophobic vs hydrophilic (and why?)
- disulfide bridge
- peptide bond
- nucleophilic addition/elimination
- polypeptides
- proteins
- hydrolysis
- proteolysis
---
- transcription
- translation
- monomer vs polymer (identify monomer for DNA, RNA, protein)
- reverse transcription
- non-coding RNA
- epigenetics
- primary structure vs secondary structure vs tertiary structure vs quatenary strucutre
- alpha helix vs parallel beta helix vs non-parallel beta helix
- how to denature a protein (4 methods)
- solvation
- Histidine
- Proline
- Glycine
- Cysteine
- protonated vs deprotonated
- secondary alpha amino group
- OIL RIG (oxidation vs reduction)
- chirality
- alliphatic